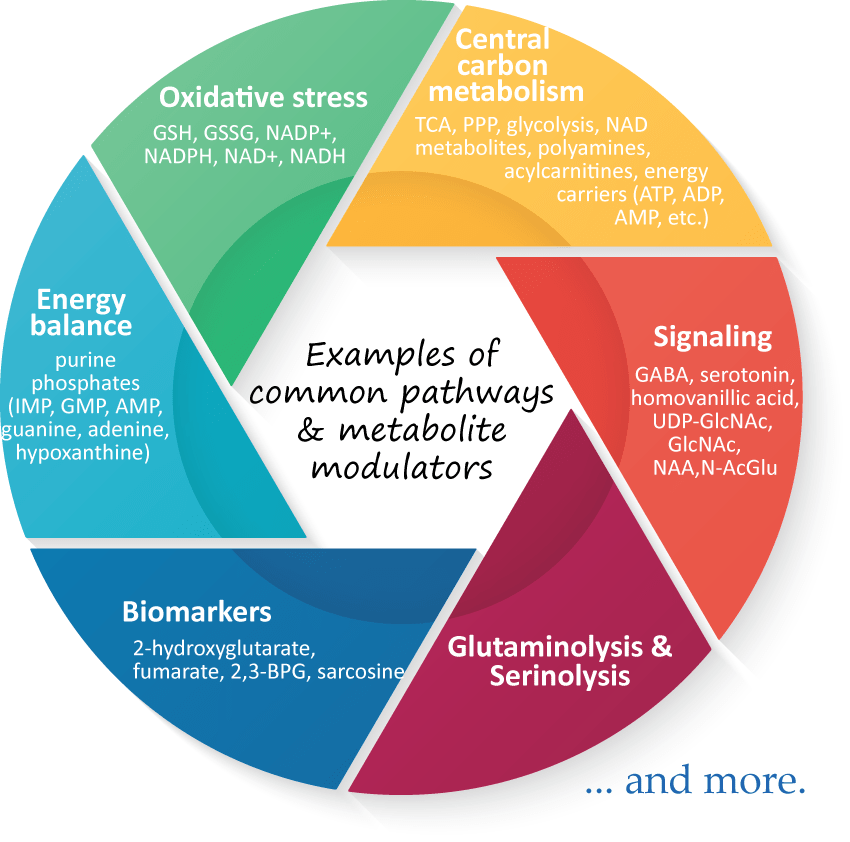
Metabolomics comprehensively characterizes small polar and lipid metabolites, yielding a snapshot of physiological processes that vary according to the pathological state of cells, tissues, and organs. Therefore, metabolomics profiling can help to identify specific metabolic changes in cancer cells and tissues, providing a deeper understanding of the role altered cellular metabolism plays in tumorigenesis, as well as in disease progression. Furthermore, targeting cancer metabolism pathways with metabolomics will be useful in determining critical biomarkers for early detection and therapeutic interventions.
Metabolomics brings us closer to the phenotype of an individual, providing a direct readout of metabolic changes that occur in the tumor microenvironment.